People are hard at work from early morning to late at night, and the amount of finished material is steadily increasing. We did lure our guests out on a little boat trip yesterday, which was very nice. Below are some photos of work in progress: discussions in the lab, identification work, drilling through mollusc shell to get to the tissue inside, piles of material and labels, and some snapshots of general life at the workshop.
Author Archives: katrine
Participant snapshots – Williams
 Name: Akanbi Bamikole Williams
Name: Akanbi Bamikole Williams
Home Institution: Nigerian Institute for Oceanography and Marine Research
What do you work with at home? I am a Principal Research Officer at my home institution with special interest in fish biology, fisheries management and benthic ecology. Presently working on marine fauna diversity in Nigeria.
What are you working on here? I have been working on taxonomic details of different families of polychaetes and some other animal groups (Molluscs and Crustatceans). I am also going through the process of preparing samples for DNA Barcoding – specimen selection, filling-in the various data sheets, taking photographs of specimens, tissues extraction and the final plate preparation. It’s been a wonderful experience.
Participant snapshots – Trond
Name: Trond R. Oskars
Home institution: University Museum of Bergen, Norway (blog here)
What do you work with at home?
I have recently been working on preparing material for the workshop by sorting benthic samples from the West-African coast, paying particular attention to the Bivalvia (clams, oysters, cockles etc.) and the Ophistobranchs (a group of gastropods/snails), as these are some of the focus groups for the workshop. I defined the specimens to morphospecies and/or genus, to ease the work for the experts who will be working on identifying them to species. I am also preparing papers on Cephalaspidea and Philinidae (snails) from my master thesis work, and preparing to start my Ph.D.
On the workshop I have been nominated as the “gastropod team”. The previous workshop got a lot of nice results on the gastropods, and it was decided to continue the work this year. My task is to organize the species by family, and pick suitable specimens from target families that we wish to do DNA barcoding on, prepare the barcoding samples, and keep the database up to date.
Aaand we’re off!
Most of our participants arrived over the course of the weekend, and work is already well under way. The 2014 workshop will focus on the Mollusca – especially the bivalves – and our ever-present polychaetes, from both the GCLME and the CCLME.
Like last year, we are based at the field station of the University of Bergen, which is a great venue for workshops.
We have – amongst our in total 15 participants – no less than nine nationalities representing nine different institutions present!
Many of us also participated in last years workshop, and it’s been very nice to have a reunion with that lovely crowd!
Some of last year’s participants had other obligations and were unfortunately unable to attend. We do have some new additions as well, and they are very welcome. As the pictures show, work is already well under way, we will get back to the strategies and results of the work in coming blog posts.
All aboard for the 2014 workshop!
Biodiversity in a dish
One of my (non-marine) colleagues asked me what I was planning on doing with the cartload of samples I was hauling into the lab – so I decided to write a little here, explaining what it is we do when we sort the samples.
The first step is of course to aquire said samples. Ours have been collected from the West African continental shelf in agreement with the Guinea Current (GCLME) and Canary Current (CCLME) Large Marine Ecosystem projects.
The sampling has been done by the R/V Dr. Fridjof Nansen.
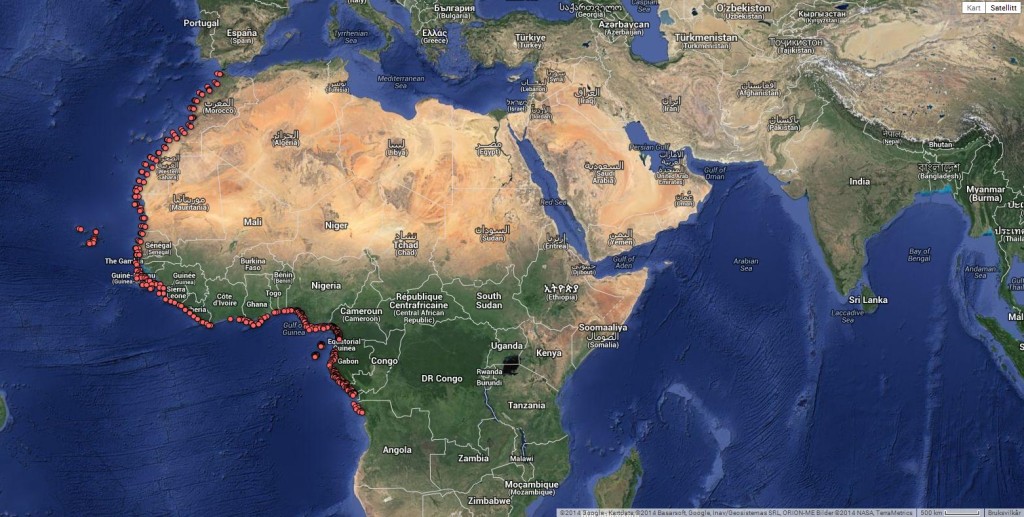
At present we house samples collected from close to 650 stations along the West African coast.
You will find all the stations and their associated metatdata in this interactive map – here’s a static version of the same:
Once the samples have arrived at the University Museum, we begin the laborious process of identifying the fauna within. The first step is – rather obviously – to separate the animals from the sediment. Whilst doing this, we also do a rough sorting of the animals to what we call the main groups: Echinoderms, Crustaceans, Polychaeta, Mollusca and so on.
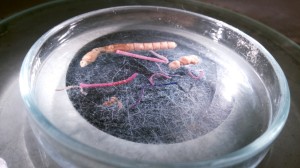
Below are some pictures from the processing of a sample collected by use of a sledge at 106 m depth in the waters of Senegal in 2011.

A 1 liter container of decanted (= the most animal rich part of the sample) sample – from the same station we will have several bottles with different fractions (decanted, 1 mm, 5 mm,…). Large or particularly conspicuos animals are picked out separately whilst processing the samples on board.

A petri dish with two tea spoons worth of unsorted sample collected by sled (this is from a decanted fraction, though – not all containers will be this intensely populated!)

At a glance..Ostracoda, Galatheidae, a Ebalia (the pink crab), isopods, amphipods and a lot of polychaeta.
Once the animals have been sorted to the main groups, they are passed on to the taxonomists, who will (do their best to) identify them to species. This is done to learn more about the rich species diversity of this region, and to compare it with the northern fauna. Some of the identified animals will be used as DNA barcode vouchers, helping us build a library of the genetic barcodes* of the marine invertebrates of western Africa.
*within the framework of the Barcode of Life Data Systems; “BOLD”
Slipper lobster
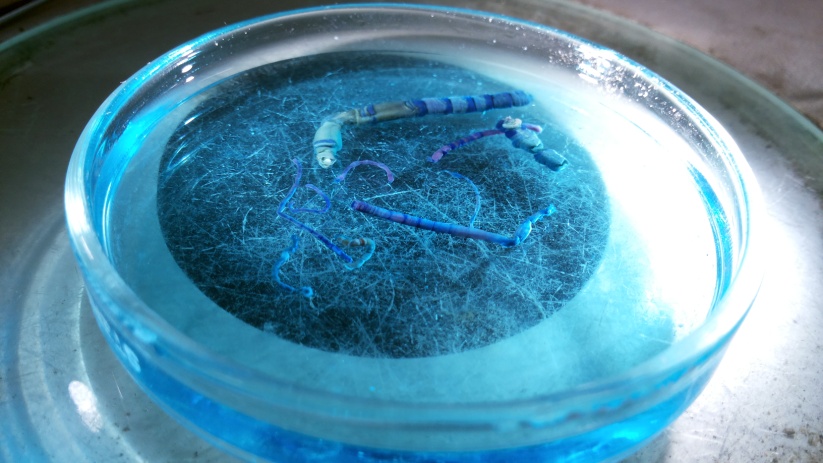
I just photographed some specimens from the family Scyllaridae, and they are such funny looking critters that I decide to share them on the blog. The Scyllaridae are found in all warm oceans and seas, and typically live from shallow water and down to depths of about 500 m (according to Wikipedia).
Pictured is a Scyllarus carpati from Mauritania, collected by sledge at 100 meters.
If you click here, you can se the distribution of the species, as well as its IUCN Red List status. We will take tissue samples from this specimen and send it for COI DNA barcoding, which will be incorporated in the BOLD database. There are records of specimens from the same genus recorded in BOLD already, but none of this particular species, as you can see if you search for Scyllarus carpati here.
Thank you to all our workshop participants!
Snapshots
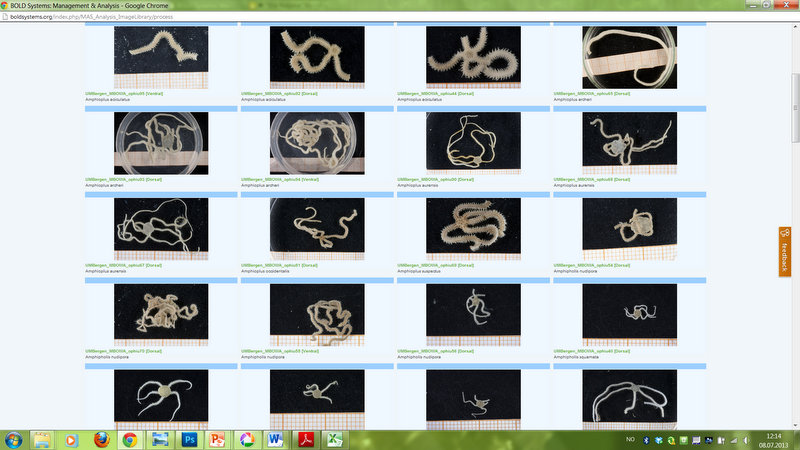
Tools of the trade – the BOLD database
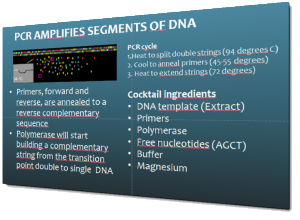
One of the topics covered in depth during the workshop is the selection of specimens and the preparation of tissue-samples. These will to be sent for DNA sequencing, and the genetic sequence will then be included in the Barcode of Life Data Systems (BOLD). The aim is to obtain standardized genetic sequences (“barcodes”) for the various taxa that we are working on. The barcode consists of a segment of approximately 650 base pairs of the mitochondrial gene cytochrome oxidase c subunit 1 (COI). You can read more about DNA barcoding on WIKIPEDIA.
There is a very real challenge connected with estimating biodiversity when many of the species are still undescribed, as is the case with many invertebrate species, especially the more obscure and diminutive groups. In such cases, barcoding can serve as a tool in screening for biodiversity, and aid the taxonomists in identifying areas where the taxonomic resolution is low.
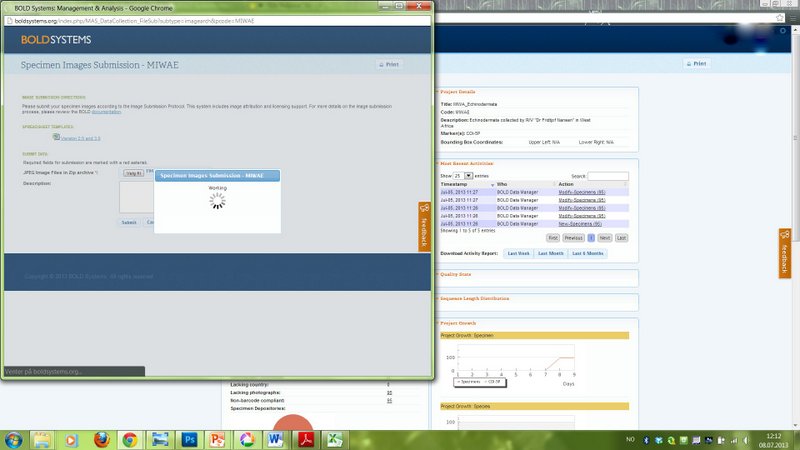
We have not yet received any barcodes for our MIWA project, but the project page on BOLD is getting populated with images and geographical information.
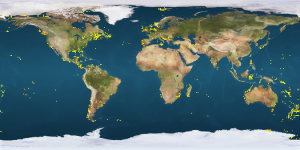
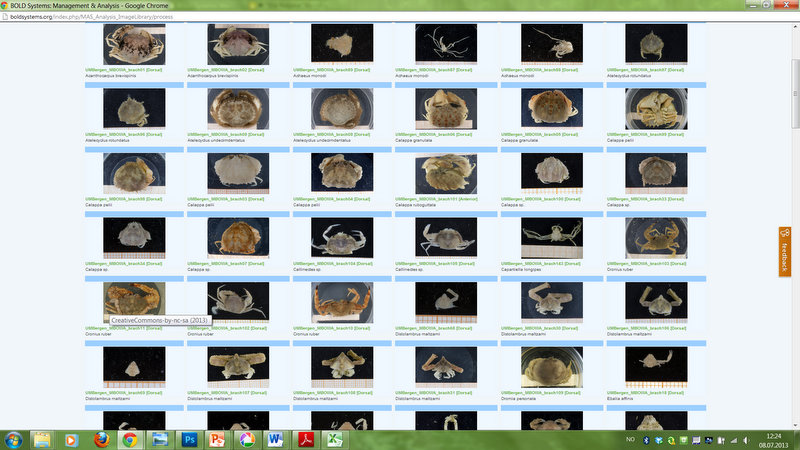
This picture shows a plot of crab species that have been DNA barcoded around the World. Notice the lack of records from West African waters.
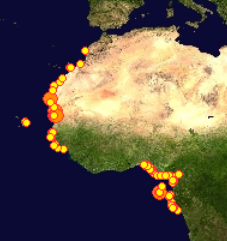
This picture shows how we are presently about to add records of about 60 crab species to the BOLD database.