Yesterday, Endre and I attended an event where the different units of the University of Bergen were invited to “..present ongoing digitalisation projects, tools and methods, and future digital solutions.”
We brought with us a poster titled “Data con carne – sources of new knowledge on biodiversity” (in Norwegian), where we presented how our barcoding efforts on both African and Norwegian material are parts of a global undertaking of building a “library of life”, and how using huge databases such as BOLD can help us gain better understanding of biodiversity – and on where to focus our efforts in unraveling the taxonomy.

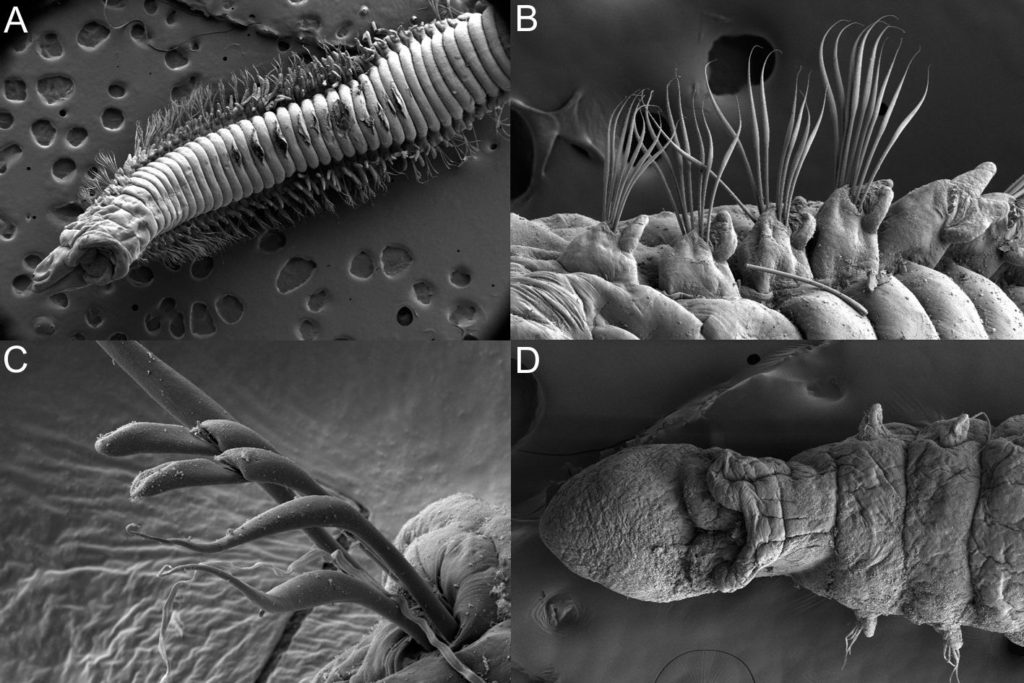
There is a very real challenge connected with estimating biodiversity when many of the species are still undescribed, as is the case with many invertebrates, especially the more obscure and diminutive groups. In such cases, barcoding can serve as a tool in screening for biodiversity, and aid the taxonomists in identifying areas where the taxonomic resolution is poor.
There is a global effort underway to establish a library of short, species specific genetic sequences. These standardized genetic sequences (“barcodes”) consists of a segment of approximately 650 base pairs of the mitochondrial gene cytochrome oxidase c subunit 1 (COI). You can read more about DNA barcoding on WIKIPEDIA.
For MIWA, we have submitted over 2700 tissue samples from over 600 morphospecies for DNA barcode sequencing through the BOLD-pipeline, where the lab work is done in Guelph, Canada, and the data is uploaded to the Barcode of Life Data Systems (BOLD Systems) .
Out of these, 1450 sequences have been obtained (54% sequencing success rate), and these cluster into close to 550 BINs (OTUs) – suggesting that the diversity may be (much!) higher than what our preliminary investigations reveal. This is not so surprising, considering how hugely diverse and little studied the invertebrate fauna of the GCLME and the CCLME is.
Below is an interactive map of the submitted samples, you can click on the stations to see the specimens that have been sent from that location, and whether or not they have gotten a barcode. You can also follow this link to find the map
What we find is that it is crucial to be able to go back and reexamine the material post-barcoding, and that is where the “con carne” part of our poster came from;
Our barcoding revealed several examples of mismatching taxonomic identifications amongst researchers in different labs and institutions in different countries. This illustrates a major challenge, as it has been shown that when benthic fauna is used in standardized methods for quality assessment and monitoring, mismatching identifications produced by different identifiers can have strong effects on indexing and valuation of ecological conditions in the same habitat.
For DNA barcoding to be useful, it is absolutely neccessary that the correlation between species name, specimen, and barcode is correct (or at least as close to it as we can get, if the species is undescribed).
We must first build this reference library before barcoding can be reliably used for identification of unknown organisms. Hence it is imperative that the voucher specimens that correspond to the genetic barcodes are deposited in a museum where it will be preserved and made available for research.
-Endre & Katrine