Kate, from Amgueddfa Cymru – National Museum Wales, has been back visiting us, and is giving us an update on how the work on the magelonid project is coming along.
The shovelhead worms
– taxonomy of magelonid polychaetes – further news
11th – 22nd February 2019
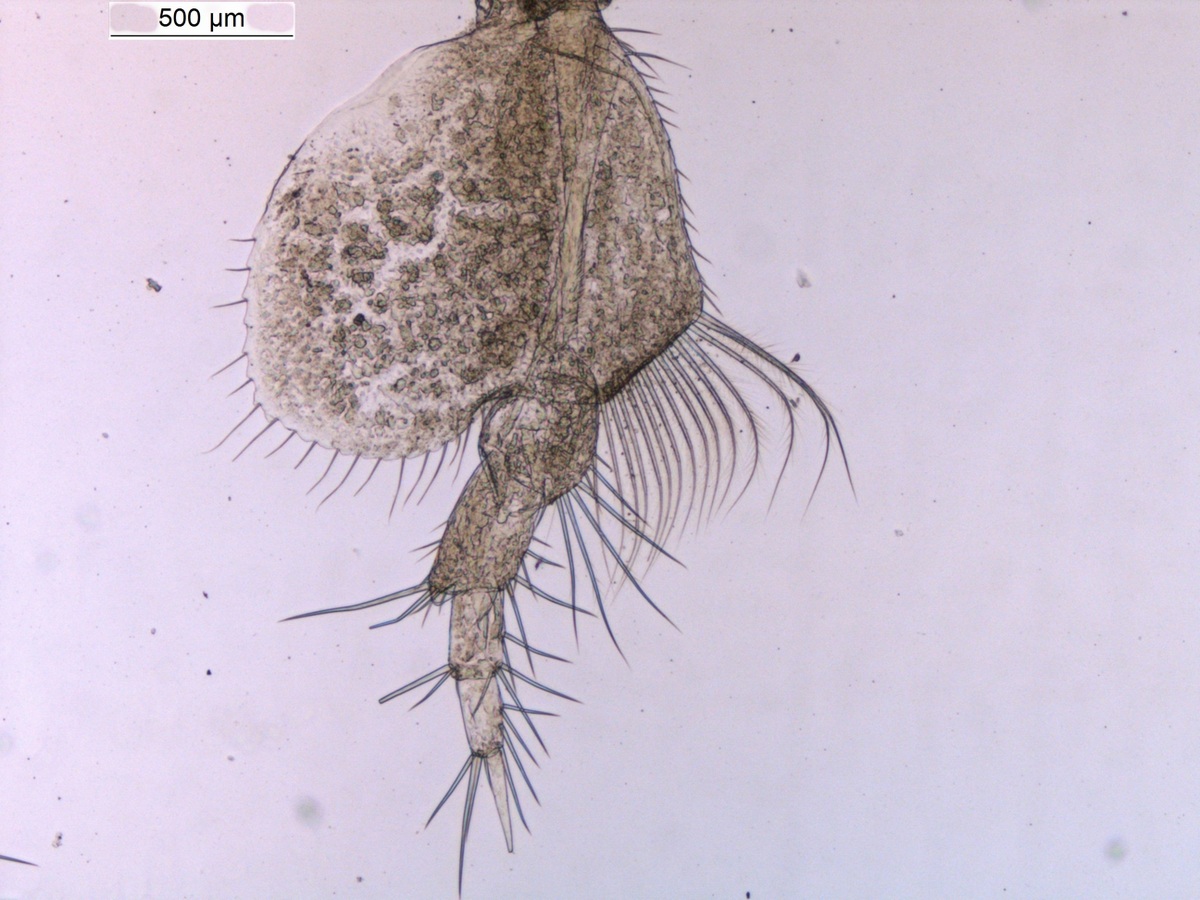
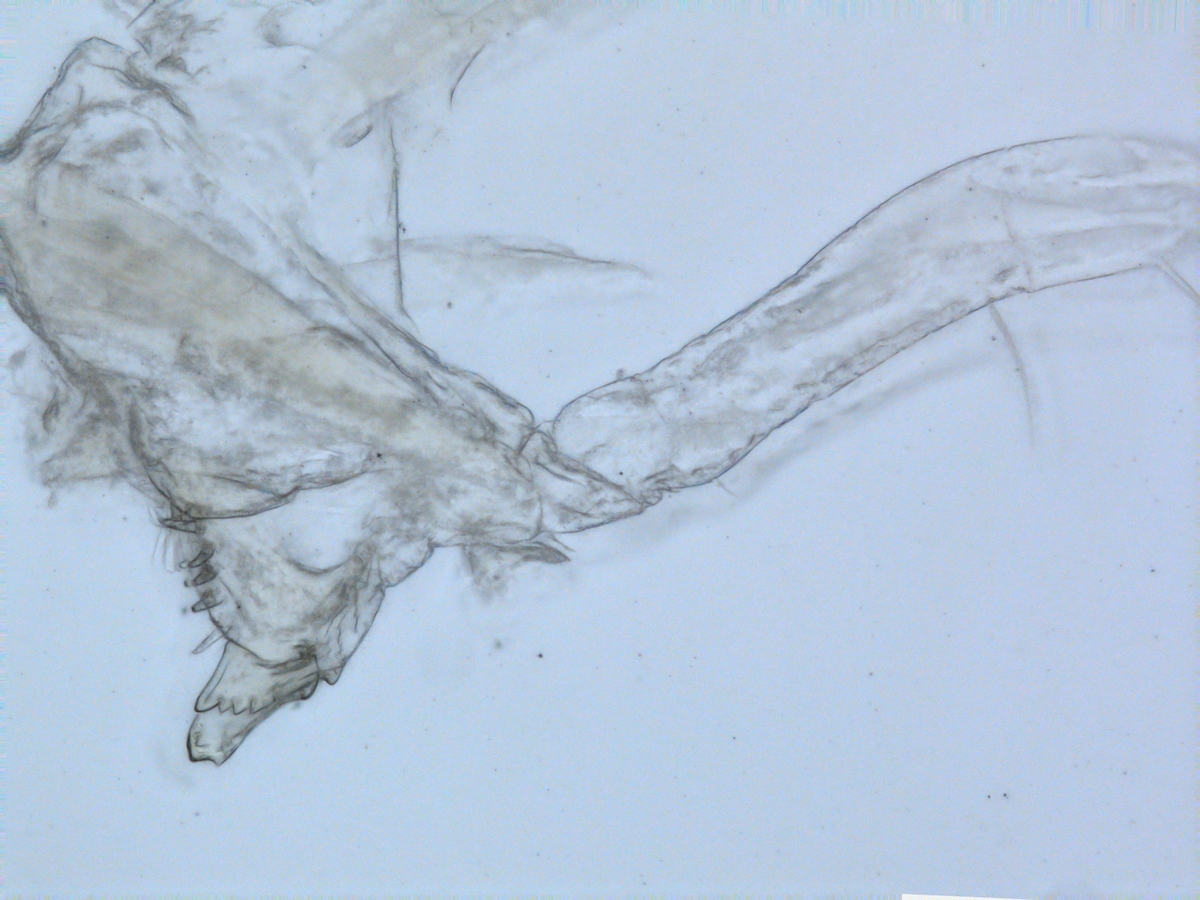
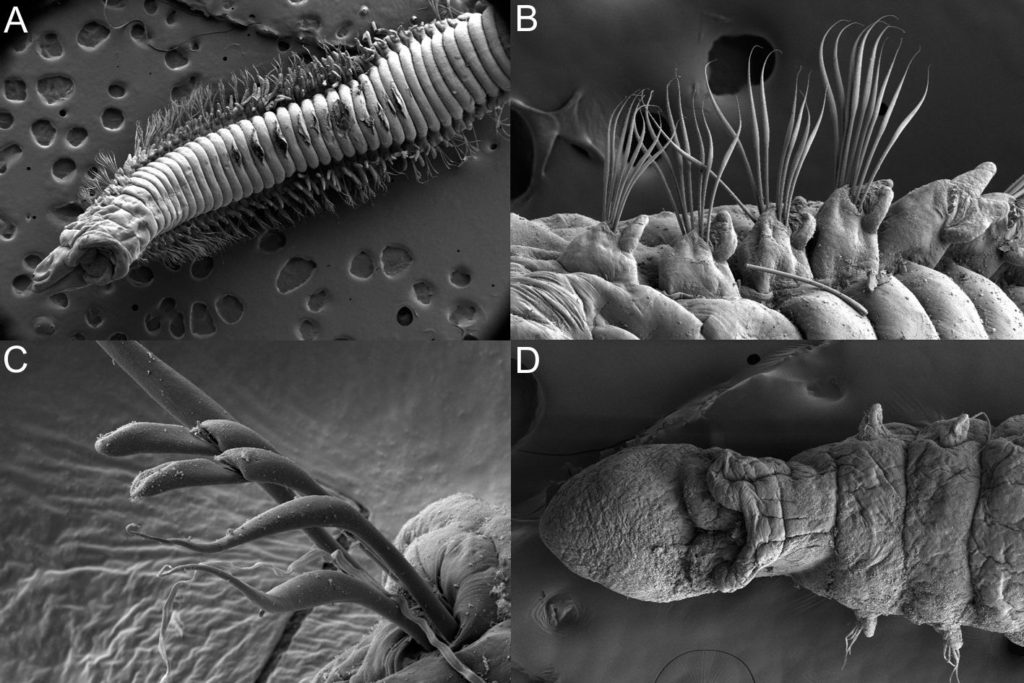
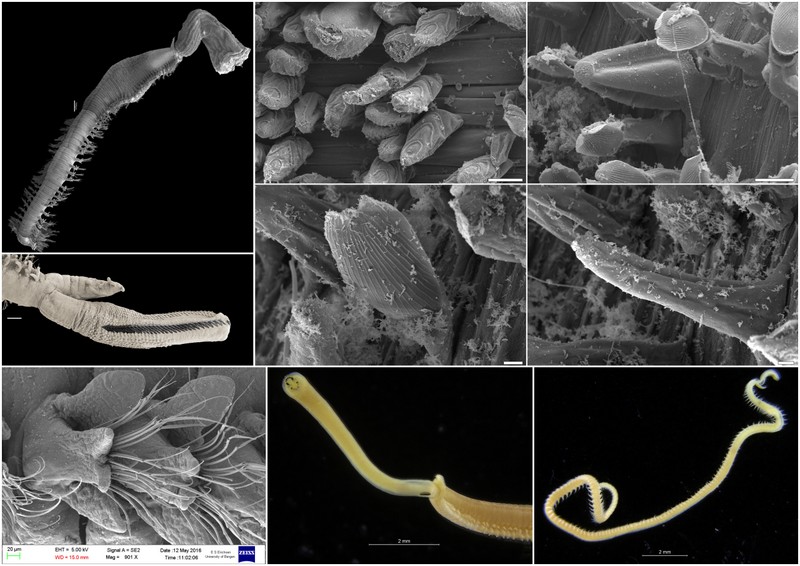
For those not familiar with magelonids, they are magnificent marine worms with shovel-shaped head regions which are utilised to dig in soft sediments. There are approximately 72 described species (many of which live in shallow water up to several hundred metres deep. Although there are magelonid species described from many parts of the world there are several regions which we know relatively little about; those of the western coast of Africa virtually nothing. With that in mind I started investigating the magelonids from the MIWA project back in 2013 and made my first visit to the University Museum of Bergen to look at the material in November 2015 and a second trip in January 2017.
Where magelonids are present you often find a huge diversity of species, with 6-10 species within a region not uncommon. However, the diversity of magelonids off the western coast of Africa has been extensive and we believe that there are in excess of 20 species within the sampling area (Morocco-Angola).
So, we have been working our way through the extensive collection of material from MIWA, identifying, describing, drawing, photographing and cataloguing all the shovelhead worm species. Alongside morphological investigations, we have taken many samples for DNA barcoding (CO1, 16S and 28S), thus allowing us to produce phylogenetic trees. With so many species and specimens to deal with, we felt it prudent to publish the results in a series of papers. To start the process, we decided to concentrate on the species that carry pigmentation in the posterior thorax. These are generally stout, short species, with short ‘frilly’ palps, and are known to construct distinct sediment tubes. One species within this group is Magelona cincta Ehlers, 1908 first described from South Africa. Normally, within a region you generally only find one or two species carrying these distinct pigment bands. However, we have found six species, five of which are new to science. Since my last visit all six of these species have been described and illustrated and the first paper on MIWA magelonids is complete and will be submitted in due course. Phylogenetic analysis has shown this group to be monophyletic.
For the next paper we are focusing on a group of magelonids that have horns on the top of their head region. These may be crenulated with distinct triangular points along the anterior margin, or may be entirely smooth. Similar species are Magelona crenulifrons Gallardo, 1968 (from Viet Nam), Magelona cornuta Wesenberg-Lund, 1949 (from the Gulf of Iran), and Magelona lusitanica Mortimer, Gil & Fiege, 2011 (from Portugal). There are five species within the MIWA material, three of which are fairly common but two of which we have only a few specimens of. The focus of this trip is to look for further specimens of those species that we have limited material of, which will help us to provide more fuller descriptions of the species.
So, we hope that by the end of my time here at UiB, we will have almost completed the descriptions of the ‘horned species’. The next step is to analyse more DNA samples, which will be done here in Bergen and I will return back to Cardiff in order to draw and photograph the five species ready for publication. Due to the huge diversity of species, it is a long process. However, by the time we have finished we will have a comprehensive understanding of the shovelhead fauna of Western Africa. Whilst many of the species have limited species ranges, we have seen an overlap with European faunas and some species with large distributions.
-Kate Mortimer-Jones