“Opisthobranch” gastropods from West Africa
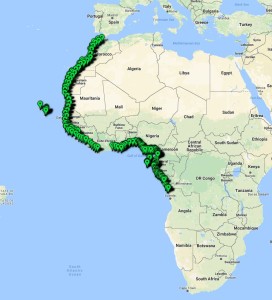
Dr Sara Castillo has recently finished her PhD (September 2017) with a thesis on the taxonomy and diversity of gastropods and bivalves from North-west Africa (Morocco to Sierra Leone), based on material collected during cruises of the Norwegian and FAO Nansen Programme and other scientific projects.
The Natural History Museum of Bergen has since 2005 collaborated in the benthic sampling of the Nansen Programme, and between 2013–16 the Museum was awarded funding from the American JRS Biodiversity Foundation to aid on the study and DNA barcode of the western African invertebrate benthic fauna. During this period Sara visited Bergen to participate in a workshop on to the taxonomy of western Africa bivalves organised by the Museum. The “opisthobranchs” gastropods (sea slugs and other related lineages) were not covered by Sara Castillo’s research and in Bergen we have so far only studied in depth the philinid snails (fam. Philinidae sensu lato).
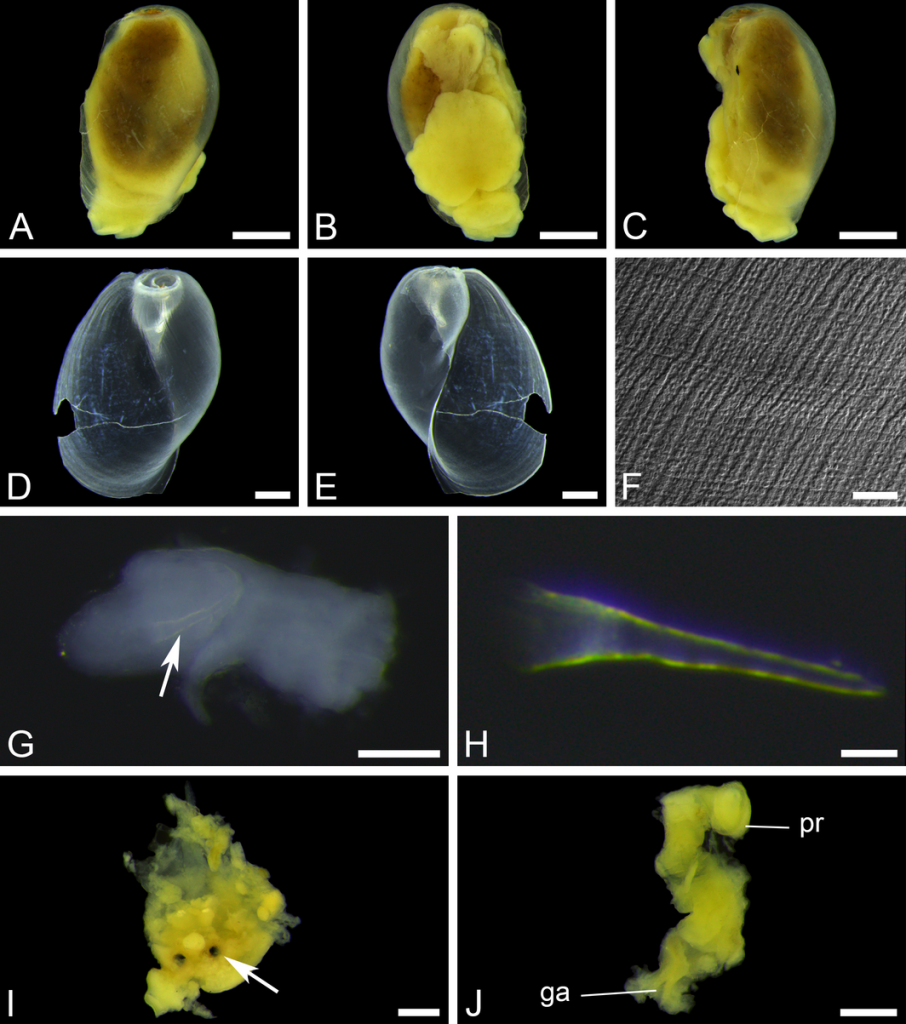
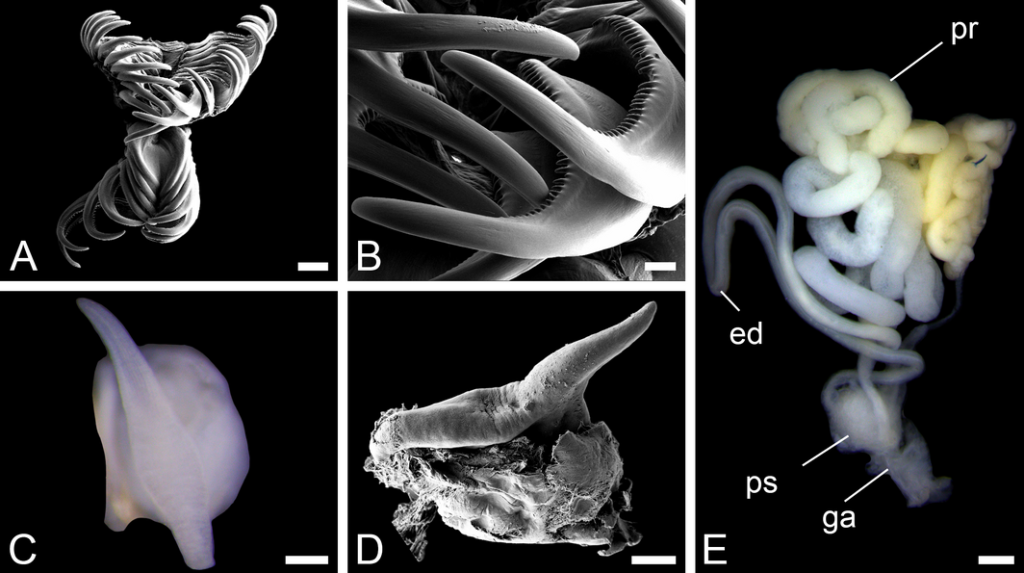
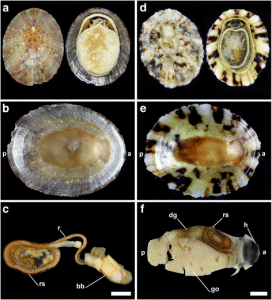
Sara is visiting the Museum for 6-weeks (1st Nov–15th Dec) to study both ours and the University of Vigo’s collection of opisthobranchs from West Africa. Representatives of all morphotypes will be DNA barcoded and identified to species level. Because of its diversity and difficult taxonomy in western Africa the nudibranch family Arminidae will receive special attention and the barcoding work will be complemented with anatomical characterization of species backed by scanning electron microscopy. Our goals are to produce an inventory of the West Africa “opisthobranchs” collected during the Nansen programme and a taxonomic review of arminid nudibranchs from the region.