Marla, a PhD student supervised by Dr Tammy Horton (NOC), Dr Andrew Gates (NOC), Dr Lawrence Hawkins (UoS), Dr Miranda Lowe (NHM) and Dr Gordon Paterson (NHM) has spent 6 weeks in the invertebrate collections at UiB.
Marla was studying the amphipods from the family Phoxocephalidae from the Western African Waters, focussing particularly on the amphipods from the sub-family Harpiniinae [crustacea; Amphipoda; Phoxocephalidae; Harpiniinae].
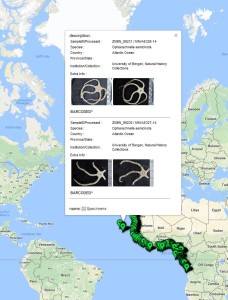
- Basuto stimpsoni, barcoded
- Harpiniopsis sp, barcoded
- Metaphoxus sp., barcoded
Phoxocephalid amphipods are highly speciose and abundant in deep sea sediments globally. Species identity is critical to understanding mechanisms driving observed biodiversity patterns and to asses community change. The aim of the project while in Bergen, was to use both DNA barcoding and traditional morphological taxonomic approaches in order to create a robust library of Phoxocephalidae species from the poorly known West African waters. Large scale projects such as Marine Invertebrates of West Africa (MIWA) provide the perfect opportunity for collaborative work.
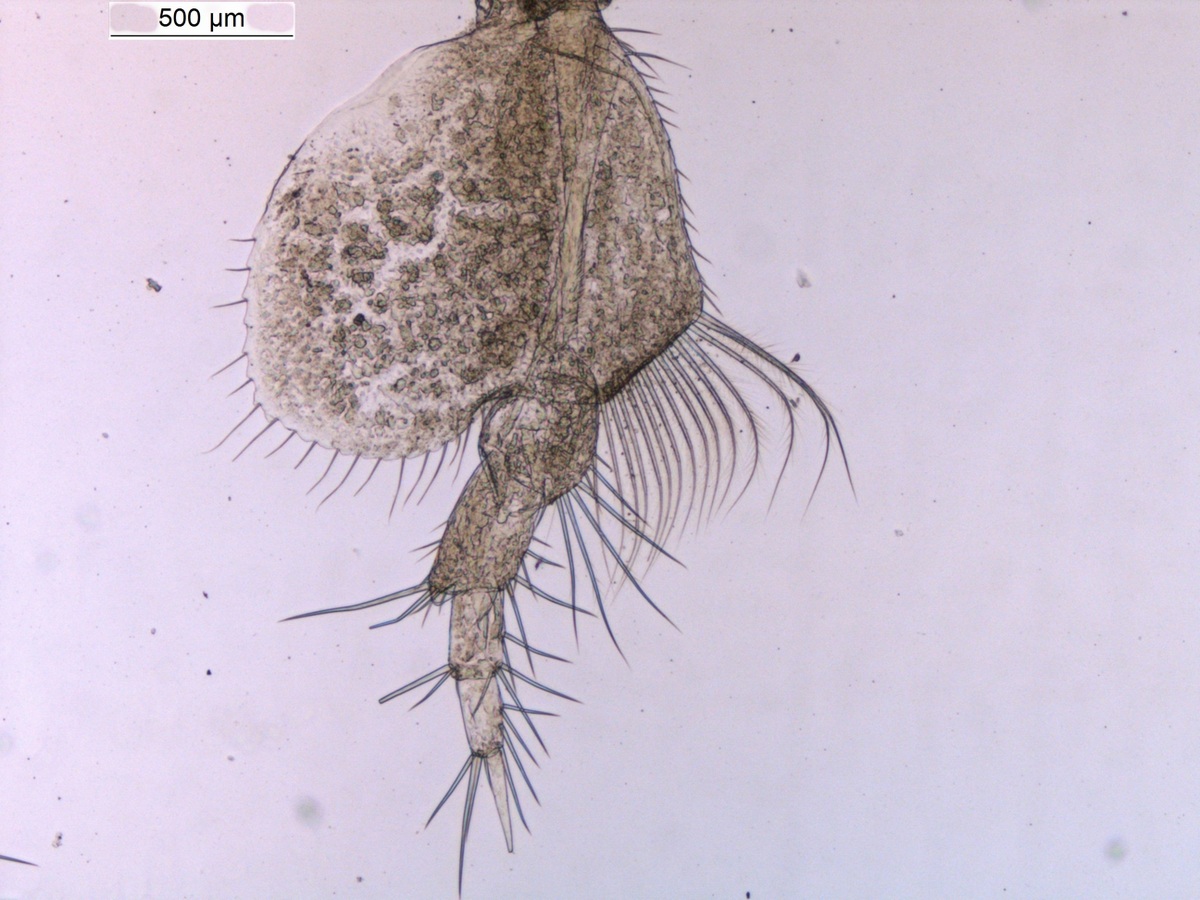
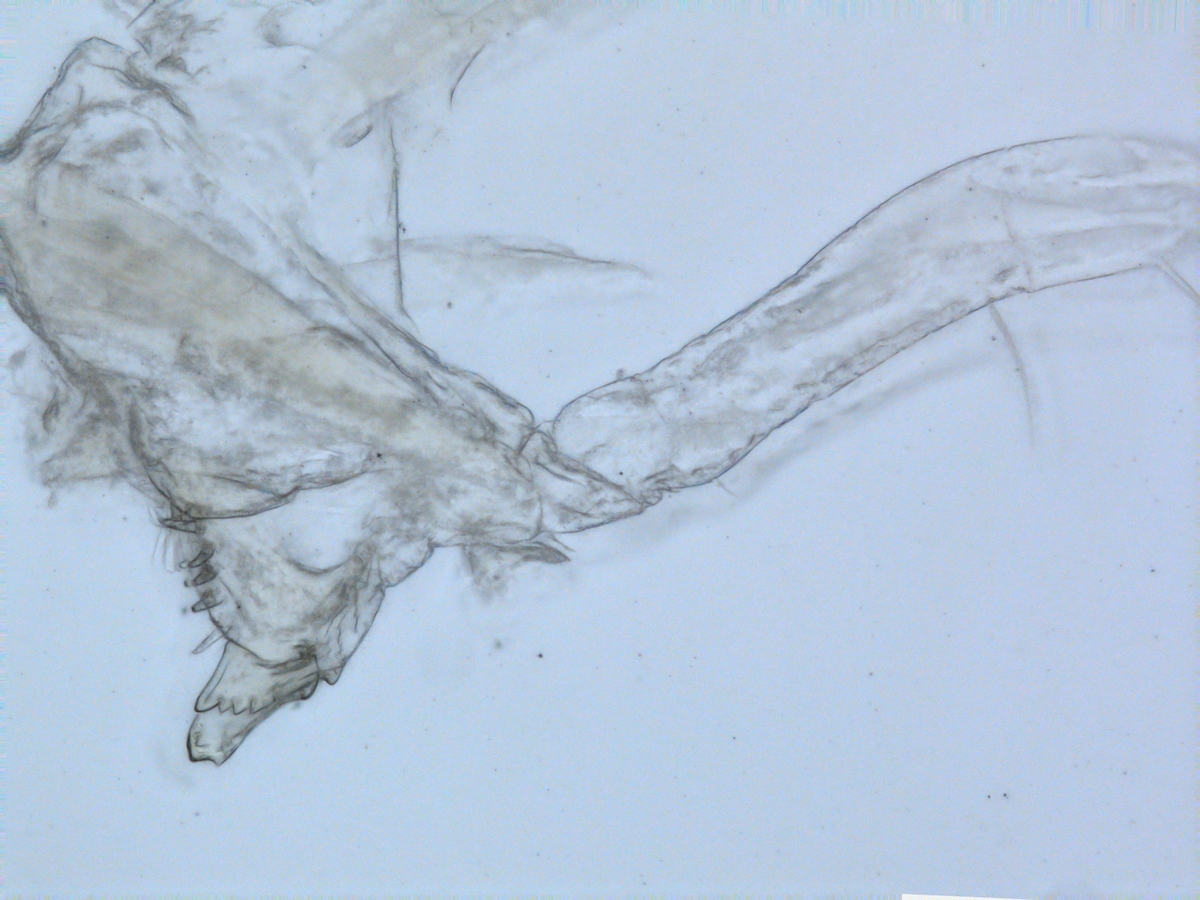
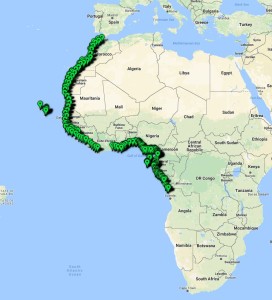
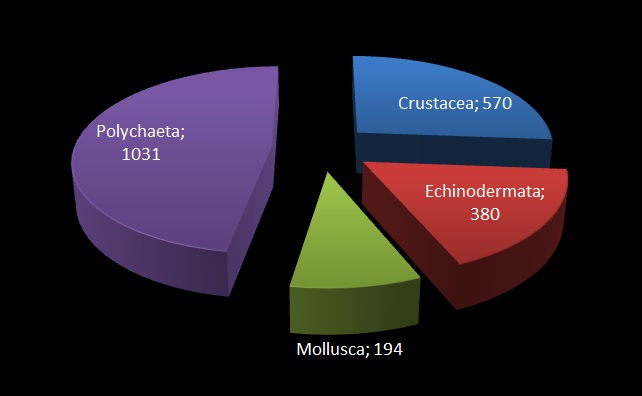
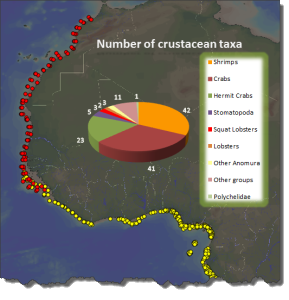
The MIWA project submitted over 2700 tissue samples from over 600 morphospecies for DNA barcode sequencing, including Crustaceans, Echinoderms, Molluscs and Polychaetes. Out of these, 45 samples were from the family Phoxocephalidae, the target taxa. Working with Dr Anne-Helene Tandberg and Prof Endre Willassen, the sequenced MIWA Phoxcephalid voucher specimens were dissected and mounted as permanent microscope slides to morphologically score them. Later, the phylogenetic analysis based on molecular and morphological characters will be compared. Each appendage was photographed on the modular (Leica CTR6000) microscope and the images were stacked, resulting in incredible photos!
Out of the 2700 tissue samples, a total of 1450 sequences were obtained (54% sequencing success rate). This is not uncommon as the ‘Universal’ barcode protocol often needs to be adapted for different taxon groups.
Working with Anne Helene within the molecular biology labs at the University of Bergen, currently developing taxon specific primers and PCR conditions for the Harpiniinae MIWA specimens which were not successfully sequenced with the Universal primers. As a starting point, an additional 13 MIWA specimens had tissue extracted for DNA, then dissected and permanent slides were made in order to morphologically score them. Each appendage was photographed and the images stacked. At this time the primers and PCR conditions are a work in progress, but we will keep you posted. However, this was a very successful trip resulting in a lot of data to analyse!
-Marla