Kate, from Amgueddfa Cymru – National Museum Wales, has been back visiting us, and is giving us an update on how the work on the magelonid project is coming along
The shovelhead worms
– taxonomy of magelonid polychaetes – an update
16th – 27th January 2017
I came to the University Museum of Bergen (UMB) back in November 2015 to work on shovelhead worms (Magelonidae) collected as part of the MIWA-project. In particular, the priority was to select specimens for DNA sequencing from each of the putative species that had been identified whilst studying the material back at the National Museum Wales in Cardiff.
The initial results came back with sequences from 45 of the 72 tissue samples that were taken, mostly from the MIWA project but also European magelonid samples for direct comparison. The corresponding tree showed some interesting results and included sequences from 13 of the 20 putative species that had been highlighted previously. However, sequences from the other species identified, and further sequences from those species that only had one or two were still needed.
So work continued back in Wales, looking for further specimens for sequencing. However, it was felt that it would be beneficial to come back to Bergen in order to further study the DNA voucher specimens that had sequences already, comparing them morphologically to new material selected. It was also hoped to select alternative specimens for sequencing which had previously failed.
So, I travelled back to UMB in January 2017, just in time to catch the end of the snow, before the rains came again to spend two weeks studying MIWA material. Each of the DNA Voucher specimens were studied in great detail, making detailed notes, drawings and full taxonomic descriptions of each.
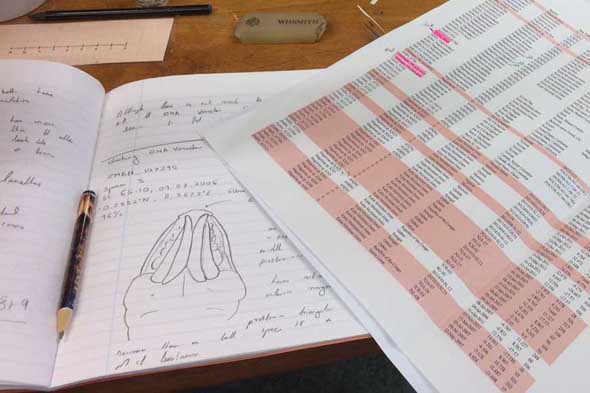
Each of the DNA voucher specimens was carefully studied taking detailed notes and making drawings of each
This enabled the selection of further specimens of the same species for sequencing but also highlighted specimens that showed differences. Consequently 74 additional specimens have now been chosen for sequencing. These have all been photographed and are now ready for tissue sampling before being sent off to Canada for sequencing.
- Work area
- Samples selected for DNA barcoding
- Katrine and Kate imaging the DNA voucher specimens
Whilst those samples are being sequenced, the process of drawing, imagining, measuring and describing each species will begin back in Cardiff with formalin fixed samples.
With so many potential new species, this could be quite a lengthy process but luckily some of this work has already started. Work commenced looking at the more stout species within samples. These species usually posses a pigment band in the posterior thorax and unlike most magelonids are known to build sediment tubes. Until now only one species with a pigment band has been described from African waters, Magelona cincta, Ehlers, 1908 from Algoa Bay, South Africa. However, a further five species with pigment bands have been found in the MIWA material, four of which are believed to be new to science. Full taxonomic descriptions and a key to these species have already been produced and the process of imaging and drawing these will begin in Cardiff next week.
Now the process of waiting for the sequences begins and wondering if it will throw any surprises into the ring. So back to Cardiff armed with two notebooks full of notes and drawings (and three less pencils!) ready to being drawing and describing.
-Kate
Thank you for visiting (always a pleasure!), and best of luck with the myriad of new species!